AI/ML for Drug Discovery and Precision Oncology
Translating variant interpretation into therapeutic opportunity
Background
A persistent challenge in precision oncology is that most cancer patients carry rare variants of unknown significance (VUS). These mutations are often overlooked in drug development because they lack frequency-based statistical power. At the same time, common oncogenic variants (e.g., in PIK3CA, TP53) have well-established drug associations that drive current therapeutic strategies.
AI/ML provides a framework to bridge this gap:
Leverage clusters of common variants to define functional and drug-response signatures.
Repurpose those signatures to annotate and prioritize rare variants that share structural or pathway-level similarity.
Enable scalable variant-to-drug mapping, ensuring that even low-frequency mutations can be connected to therapeutic opportunities.
Phase 1: AI-Driven Variant Annotation (complete)
Problem: Rare variants lack annotation, limiting clinical actionability.
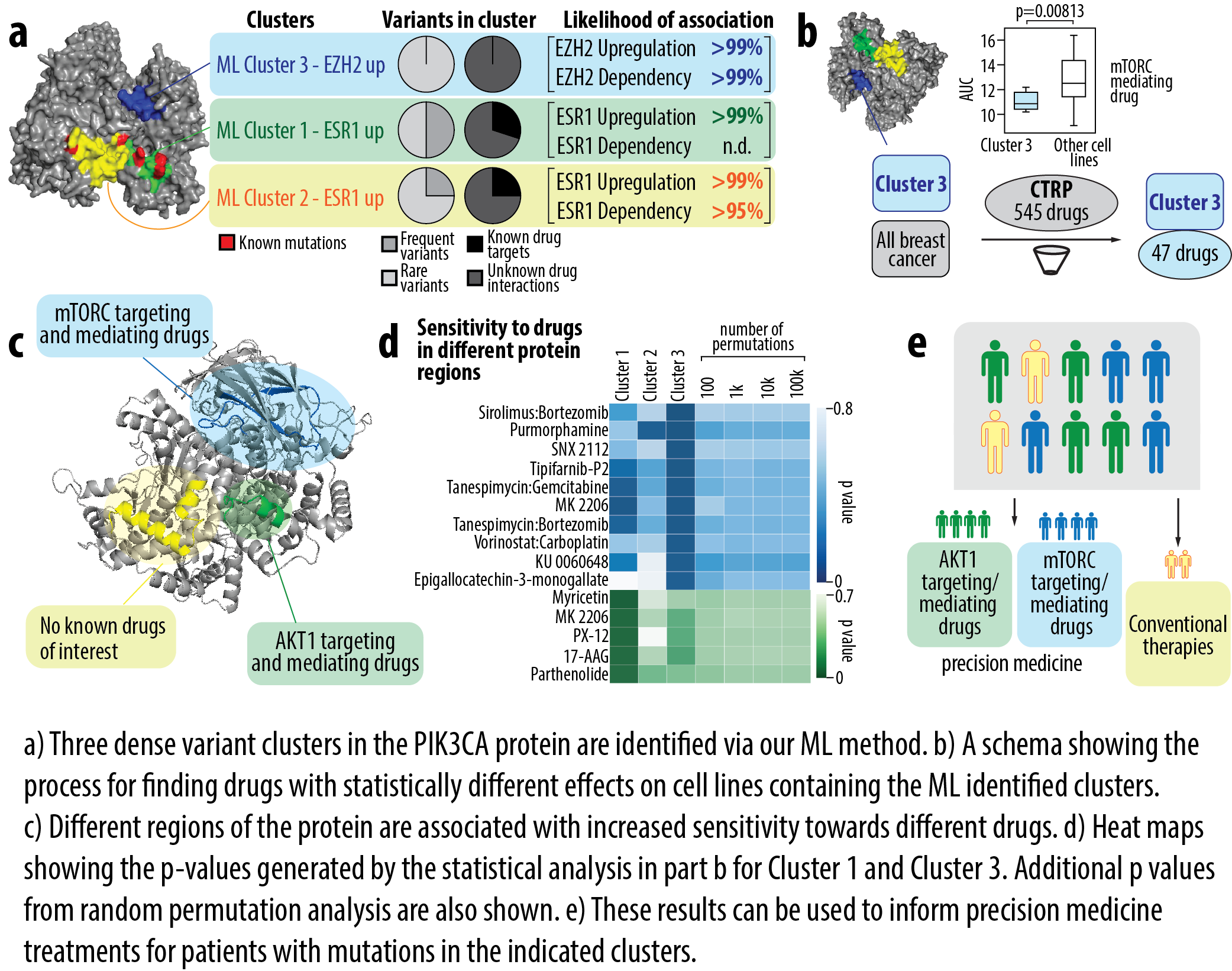
Approach: Developed AI/ML pipelines to cluster variants in 3D protein space and link them to phenotypic readouts such as ESR1/EZH2 pathway activity.
Methods: Density-based clustering, Random Forest
Data: Cancer cell line data from DepMap/CCLE, Variant annotation data from ClinVar
Outcome:
Identified common variant clusters (e.g., PIK3CA hotspots) enriched for sensitivity to mTORC and AKT inhibitors.
Repurposed these associations to annotate rare variants mapping to the same clusters.
Highlighted TP53 clusters with opposing effects on ESR1 signaling, suggesting divergent therapeutic responses.
Phase 2: AI/ML Pipelines for Drug Response Prediction (complete)
Idea: Common variant clusters can act as templates for drug-response phenotypes, extending therapeutic predictions to rare variants.
Approach: Built supervised ML pipelines trained on pharmacogenomics datasets to generalize drug-response predictions from common → rare clusters.
Methods: Density-based clustering, Random Forest, XGBoost, Graph Neural Networks
Data: Pharacogenomics datasets like the Cancer Therapeutics Reserch Portal (CTRP) and Genomics of Drug Sensitivity in Cancer (GDSC)
Outcome:
Annotated >12,000 variants across breast cancer datasets.
Connected rare variants to existing targeted therapies via shared functional clusters.
This work has the implication to expand the precision oncology therapies available to patients by 25%